Scientific understanding of the human genome has made great strides over the years but much remains unknown, particularly, with regard to gene function at the molecular and cellular level. The Molecular Phenotypes of Nulle Alleles in Cells (MorPhiC) Consortium aims to fill this gap by compiling a catalogue of gene variant characteristics. An overview of the Consortium’s goals, deliverables, and implementation plans was recently published in Nature.
“Understanding gene functions across the genome is key to revealing the role that the genes play and how they interact with each other in biological pathways and networks,” said Ali Shojaie, a professor of biostatistics at the University of Washington School of Public Health who is a member the MorPhiC Consortium, specifically, the data analysis and validation center located at the Fred Hutch Cancer Center.
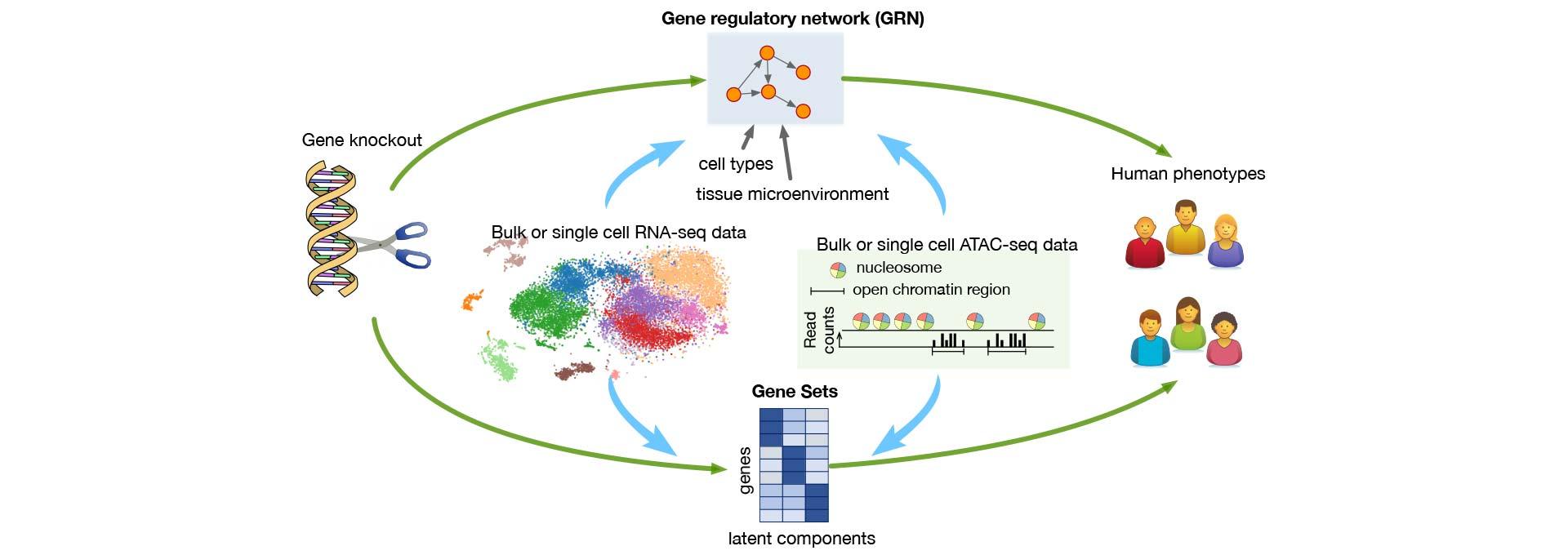
“Studying the effects of turning off each human gene would provide valuable insights into their functions and help reveal how genes influence each other and various phenotypes.”
In its paper, MorpPhiC noted that what is known about the molecular and cellular function of human genes comes from studies of specific conditions, which has resulted in an imperfect understanding of genes that is biased toward certain diseases such as cancer.
To mitigate this bias, a systematic effort is needed to create a robust catalogue of human gene functions. To accomplish this, MorPhiC’s initial phase aims to:
- Deliver a catalogue of cellular and molecular phenotypes of 1,000 protein-coding genes
- Optimize and scale up null generation strategies
- Compare a core set of phenotypes using various null-allele generation technologies
- Identify the main barriers to genome-scale null-allele phenotype characterization, including assay universality and scalability
- Develop the analysis and visualization tools to facilitate the dissemination and utilization of MorPhiC data
- Identify approaches that will enable scaling to the remaining protein-coding genes through robust, reliable and cost-effective methods
- Integrate the phenotypic data with existing knowledge and make it available to the public
The MorPhiC Consortium currently consists of four data production centers (DPCs), one data resource and administrative coordinating center (DRACC) and three data analysis and validation centers (DAVs). MorPhiC will provide open access to its data for use in future gene studies.
Fred Hutch researchers and affiliate UW faculty members Wei Sun are also PIs of the FHCC-UW data analysis and validation center.